Commits on Source (28)
-
Kien Le authored6ce60555
-
Lan Dam authored1dce2cbb
-
Lan Dam authoredf6c0cf2f
-
Lan Dam authored79a170f3
-
Lan Dam authored9cb43683
-
Lan Dam authoredc448b120
-
Lan Dam authored2f175f62
-
Lan Dam authored657e0ea4
-
Lan Dam authored42572c95
-
Lan Dam authoredd50145d1
-
Lan Dam authored1929e3ac
-
Lan Dam authored6484abba
-
Lan Dam authored88e53586
Showing
- documentation/01 _ Table of Contents.help.md 13 additions, 7 deletionsdocumentation/01 _ Table of Contents.help.md
- documentation/04 _ Search List of Directories.help.md 0 additions, 0 deletionsdocumentation/04 _ Search List of Directories.help.md
- documentation/05 _ Read from Data Card.help.md 0 additions, 0 deletionsdocumentation/05 _ Read from Data Card.help.md
- documentation/06 _ Select SOH.help.md 0 additions, 0 deletionsdocumentation/06 _ Select SOH.help.md
- documentation/07 _ Select Mass Position.help.md 0 additions, 0 deletionsdocumentation/07 _ Select Mass Position.help.md
- documentation/08 _ Select Waveforms.help.md 7 additions, 5 deletionsdocumentation/08 _ Select Waveforms.help.md
- documentation/09 _ Gap Display.help.md 0 additions, 0 deletionsdocumentation/09 _ Gap Display.help.md
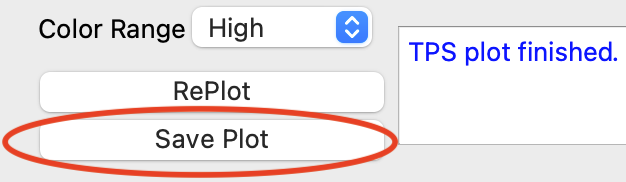
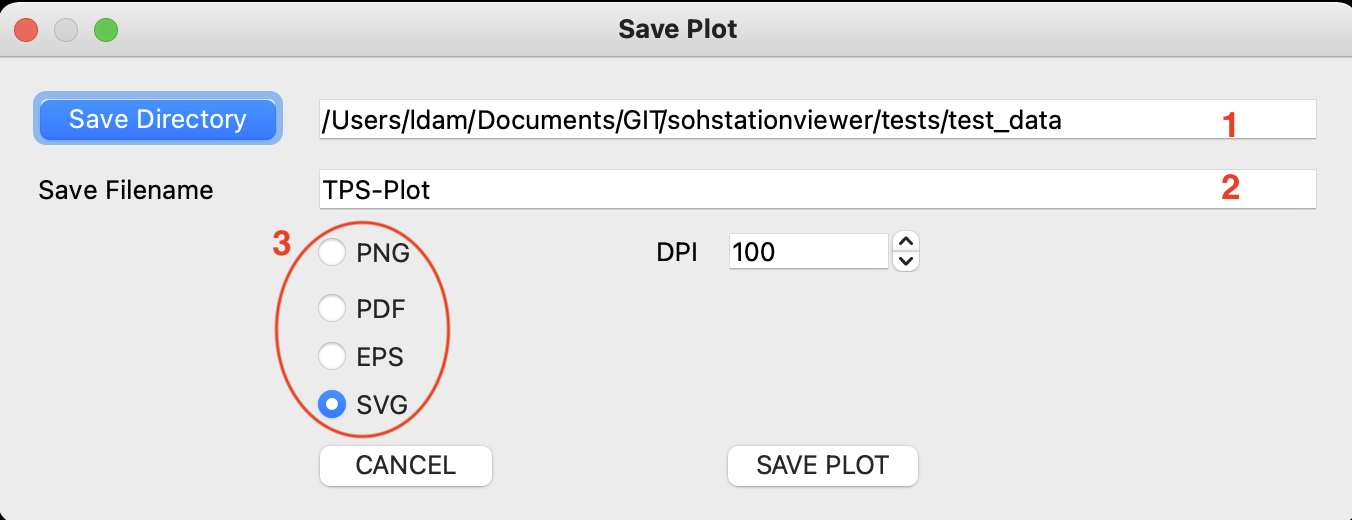
- documentation/11 _ Save Plots.help.md 60 additions, 0 deletionsdocumentation/11 _ Save Plots.help.md
- documentation/12 _ Search SOH n LOG.help.md 0 additions, 0 deletionsdocumentation/12 _ Search SOH n LOG.help.md
- documentation/99 _ test.md 1 addition, 1 deletiondocumentation/99 _ test.md
- documentation/images/save_plots/question_on_changing_black_mode.png 0 additions, 0 deletions...ion/images/save_plots/question_on_changing_black_mode.png
- documentation/images/save_plots/save_button_soh.png 0 additions, 0 deletionsdocumentation/images/save_plots/save_button_soh.png
- documentation/images/save_plots/save_button_tps.png 0 additions, 0 deletionsdocumentation/images/save_plots/save_button_tps.png
- documentation/images/save_plots/save_button_wf.png 0 additions, 0 deletionsdocumentation/images/save_plots/save_button_wf.png
- documentation/images/save_plots/save_file_dialog.png 0 additions, 0 deletionsdocumentation/images/save_plots/save_file_dialog.png
- documentation/img.png 0 additions, 0 deletionsdocumentation/img.png
- documentation/recreate_table_contents.png 0 additions, 0 deletionsdocumentation/recreate_table_contents.png
- sohstationviewer/conf/constants.py 4 additions, 1 deletionsohstationviewer/conf/constants.py
- sohstationviewer/controller/processing.py 86 additions, 97 deletionssohstationviewer/controller/processing.py
- sohstationviewer/controller/util.py 6 additions, 5 deletionssohstationviewer/controller/util.py
File moved
File moved
File moved
documentation/11 _ Save Plots.help.md
0 → 100644
File moved
39.6 KiB
9.79 KiB
31.1 KiB
13.8 KiB
78.8 KiB
documentation/img.png
deleted
100644 → 0
3.19 KiB
documentation/recreate_table_contents.png
0 → 100644
25.7 KiB